 |
|
|
|
 |
|
|
 |

|
 |
 |
 |
 |
 |
 |
 |
Single Molecule Biochemistry |
Because our nanocircuits are sensitive to chemistry at the single molecule scale, they can be used
to investigate complex molecules like proteins and enzymes and uncover new information about these
molecules' chemical activity and binding partners.
Exploring chemistry at the single molecule scale is a fascinating possibility and in the past few
years, many scientists have adopted Förster resonance energy transfer (FRET), a fluorescence
technique for watching the motions of individual proteins. However, fluorescence limitations like
bleaching and finite fluorophore brightness limit FRET from seeing fast dynamic motions or doing
long-term measurements on a single molecule.
The nanocircuit technique improves on those limits by providing microsecond resolution over unlimited
durations. Long, high-resolution recordings from a single molecule allow us to understand the
components of an enzyme's kinetics, to see short-lived, intermediate states, and to find rare,
anomalous events that can be the sympotom or the trigger for malfunction and disease.
So far, we have uncovered new information about three enzymes: lysozyme, the Klenow fragment of
DNA polymerase I, and protein kinase A. The publications below describe the specific findings with
each, and describe our efforts to generalize the technique to virtually any protein of interest. You
may also be interested in a
1-hour lecture on the topic given by Prof. Collins.
|
 |
Publications |
Single Molecule Bioelectronics (a book chapter reviewing all of the following publications)
Y. Choi, G.A. Weiss, & P.G. Collins, in Comprehensive Bioelectronics.
S. Carrera & K. Iniewski, Eds. (Cambridge University Press, 2013).
Single Molecule Recordings of Lysozyme Activity (review article)
Y. Choi, G.A. Weiss & P.G. Collins, Phys. Chem. Chem. Phys. 15, 14879-95 (2013).
Electronic Measurements of Single-Molecule Catalysis by cAMP-Dependent Protein Kinase A
P.C. Sims, I.S. Moody, Y. Choi, C. Dong, M. Iftikhar, B.L. Corso, O.T. Gul, P.G. Collins & G.A. Weiss,
J. Am. Chem. Soc. 135, 7861–8 (2013).
Electronic Measurements of Single-Molecule Processing by DNA polymerase I (Klenow fragment)
T.J. Olsen, Y. Choi, P.C. Sims, O.T. Gul, B.L. Corso, C. Dong, W.A. Brown, P.G. Collins & G.A. Weiss,
J. Am. Chem. Soc. 135, 7855–60 (2013).
Dissecting Single-Molecule Signal Transduction in Carbon Nanotube Circuits with Protein Engineering
Y. Choi, T.J. Olsen, P.C. Sims, I.S. Moody, B.L. Corso, M.N. Dang, G.A. Weiss & P.G. Collins,
Nano Lett. 13, 625-31 (2013).
Single Molecule Dynamics of Lysozyme Processing Distinguishes Linear and Cross-linked Peptidoglycan Substrates
Y. Choi, I.S. Moody, P.C. Sims, S.R. Hunt, B.L. Corso, D.E. Seitz, L.C. Blaszcazk, P.G. Collins & G.A. Weiss,
J. Am. Chem. Soc. 134, 2032-5 (2012).
Single-Molecule Lysozyme Dynamics Monitored by an Electronic Circuit
Y. Choi, I.S. Moody, P.C. Sims, S.R. Hunt, B.L. Corso, G.A. Weiss & P.G. Collins
Science 335, 319-24 (2012).
|
 |
Acknowledgements |
| This research is financially supported by the NSF and the NIH. |


 |
|
|
|
 |
|
|
|

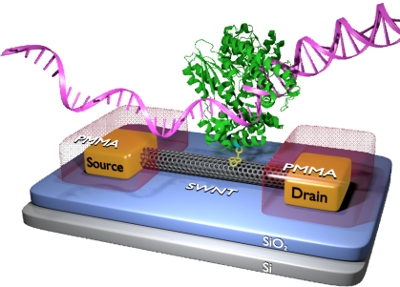
Cartoon depiction of DNA polymerase 1 creating a DNA duplex, all while tethered to a SWNT nanocircuit.

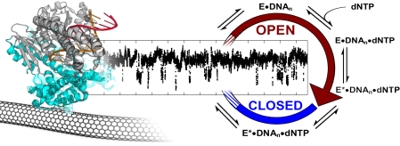
Electronic signal generated by DNA polymerase 1, overlaid with a schematic of its complex catalytic cycle.

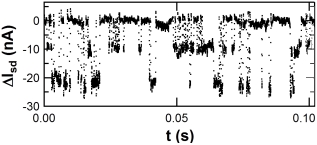
Signal generated by PKA while phosphorylating Kemptide substrate.

|
 |


