Reference Guide¶
This page documents the public function and class library for the IRATE format tool package. These modules can be used by other programs or users to simplify various processing tasks relating to the IRATE format. The following modules are present:
General tools of use with the IRATE format, such as creating standard sections or quickly extracting particles. Note that these functions should be imported directly from the irate namespace.
Code for testing if a file obeys the IRATE format and tools for extending this process (see Custom Validators/Extending the IRATE Format)
These modules all contain functions used by the Command Line Scripts and Conversion Tools to convert files in other formats from or to IRATE. This documentation is provided to allow other programs to easily script such conversions without using the command line tools.
Not that the base irate module contains most of the irate.core functionality, as well as __version__, which contains a string with the version of the IRATE tools, and formatversion, which contains an integer specifying the format version.
irate.core Module¶
The irate.core module contains the core utilities used by IRATE. These include a variety of convinience functions for working with IRATE format files. While they are technically part of the irate.core module, they are all also included in the base irate namespace, so the best way to import these is to do:
from irate import create_irate
rather than importing from irate.core.
- irate.core.add_cosmology(iratefile, omegaM=None, omegaL=None, h=None, s8=None, ns=None, omegaB=None, cosmoname=None, update=True, verbose=False)¶
Creates or updates a cosmology section of an IRATE file.
This function creates an IRATE file or opens it if it already exists, and either adds the given cosmology-related values to it, checks that they match with what’s in the existing file, or updates the cosmology with the given values.
Parameters: - iratefile – The name of the file to open or a h5py.File object.
- omegaM (float) – The present day matter density or None to skip this parameter.
- omegaL (float) – The present day dark energy density or None to skip this parameter.
- h (float) – The reduced Hubble Parameter
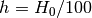 or None to skip this
parameter.
or None to skip this
parameter. - s8 (float) –
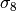 , the amplitude of the power spectrum at 8 Mpc/h, or None
to skip this parameter.
, the amplitude of the power spectrum at 8 Mpc/h, or None
to skip this parameter. - ns (float) – The index of the primordial power spectrum or None to skip this parameter.
- omegaB (float) – The present day baryon density or None to skip this parameter
- cosmoname (str) – The name of the cosmology as a string or None to skip this value. This attribute is optional in the IRATE format.
- update (bool) – If an existing file is given and this is True, the parameters will be updated with those given to this function. Otherwise, the file will be checked to ensure the file matches. If this is False and the file does not exist, an exception will be raised.
- verbose (bool) – If True, informational messages are printed as the function does various actions.
Raises: - TypeError – If the iratefile input is invalid
- ValueError – If update is False and the file’s values don’t match those passed into the function, or the file does not exist.
- KeyError – If update is False and there are values passed that aren’t in the file.
- irate.core.add_standard_cosmology(iratefile, cname, update=True, verbose=False)¶
Adds or updates a cosmology from the name of a standard cosmology.
The cname parameter gives the name of the cosmology to be used from the list below, while all other parameters are the same as for the add_cosmology() function.
‘WMAP7’
LCDM Cosmological parameters for the 7-year WMAP data (Komatsu et al. 2011). For details, see http://lambda.gsfc.nasa.gov/product/map/dr4/params/lcdm_sz_lens_wmap7.cfm
‘WMAP5’
LCDM Cosmological parameters for the 5-year WMAP data (Komatsu et al. 2009). For details, see http://lambda.gsfc.nasa.gov/product/map/dr3/params/lcdm_sz_lens_wmap7.cfm
‘WMAP3’
LCDM Cosmological parameters for the 3-year WMAP data (Spergel et al. 2007). For details, see http://lambda.gsfc.nasa.gov/product/map/dr2/params/lcdm_sz_lens_wmap7.cfm
‘WMAP1’
LCDM Cosmological parameters for the 1-year WMAP data. For details, see Spergel et al. 2003.
- irate.core.get_irate_particle_nums(fn, validate=True)¶
Convinience function to determine the number of particles of each type in an IRATE format file.
Parameters: Returns: a 3-namedtuple (ndark,nstar,ngas)
- irate.core.get_irate_catalog_halo_nums(fn, validate=True)¶
Convinience function to determine the number of entries in an IRATE halo catalog file.
Parameters: Returns: an integer with the total number of halos
- irate.core.create_irate(fn, dark, star, gas, headername, headerdict=None, compression=None)¶
A Convinience function to create an IRATE format file data as numpy arrays. This will overwrite a currently existing file.
Parameters: - fn (str) – The file name to use for the new file
- dark – The data for the dark matter particles. Should be a dict with keys ‘Position’, ‘Velocity’, ‘Mass’, (and possibly others) mapping to numpy.ndarray arrays. Alternatively, it can be a structured array (i.e. dtype with names and formats) with the names of the dtype giving the names of the resulting datasets.
- star – The data for the star particles. See dark for format.
- gas – The data for the gas particles. See dark for format.
- headername (str) – The group name to use for the header entry or None to create no header.
- headerdict – A dictionary mapping strings to attribute values. These will be used for the attributes of the header entry. If None, an attribute will be added to indicate header content is missing.
- compression – The type of compression to use for the datasets. Can be any of the compression that h5py supports on your system (‘gzip’,’lzf’, or ‘szip’) or None to do no compression.
Returns: The h5py.File of the newly-created file (still open).
Raises: - IRATEFormatError – If any of the data arrays are missing the necessary fields.
- ValueError – If the input data is invalid.
- irate.core.get_all_particles(fn, ptype, dataname, subsample=None, validate=True)¶
Convinience function to build an array with all the particles of a requested type or types (i.e. all subgroups, if present, will be visited).
Parameters: - fn (str) – The filename of an IRATE formatted file or a h5py.File object.
- ptype (str) – The particle type: ‘Dark’,’Star’, ‘Gas’, a sequence of a mix of those three, or None for all particles of all types.
- dataname (str) – The dataset name to extract (e.g. ‘Position’, ‘Mass’)
- subsample (int) – Gives the stride of the data - e.g. each array will be spliced to only return every subsample data point. If None, all the data will be returned.
- validate (bool) – If True, the file will be validate as IRATE format. If ‘strict’, strict validation will be used. Otherwise no validation will be performed.
Returns: A 3-tuple (data,grpidx,grpmap) where data is the data arrays, grpidx is an int array with the same first dimension as data, and grpmap is a dictionary mapping the values in grpidx to group names.
Raises ValueError: If a group does not have the requested dataname.
- irate.core.scatter_files(infile, outfile=None, splitgroups=False)¶
Splits a single IRATE file into separate files for each dataset.
Parameters: - infile – A file name or an h5py.File object to be scattered.
- outfile – The filename or an h5py.File object to use as the new base IRATE format file or None to overwrite the old name. Other output files will be placed in the same directory as this file.
- splitgroups (bool) – If True, also split groups into separate files.
Returns: A list of filenames that were created with the base name as the first.
- irate.core.gather_files(infile, outfile=None)¶
Combine all external file that are part of a master IRATE file into a single monolithic file.
Note
This function can be driven from the command line via the irategather script. See Command Line Scripts and Conversion Tools for details.
Parameters: - infile – A file name or an h5py.File object to be gathered.
- outfile – The filename or an h5py.File object to use as the new IRATE format file or None to overwrite the old name.
Returns: (outfilename,filelist) where outfilename is the name of the output file, and filelist is a list of all the files gathered together to create this file.
- irate.core.get_metadata(dataset, metadataname)¶
Gets a metadata value from an IRATE file dataset.
Parameters: Returns: The value of the metadata requested, or None if the metadata with the requested name does not exist.
Raises TypeError: If the dataset is not a dataset
- irate.core.set_metadata(dataset, metadataname, value, group=None)¶
Sets a metadata value for an IRATE file dataset.
Parameters: Note
Any other datasets with the same as this one below this group in the heirarchy will be assigned the same units (unless that dataset overrides them itself).
Raises: - TypeError – If dataset is not a dataset.
- ValueError – If group is not a parent of dataset.
Note
A MetadataHiddenWarning will be issued as a warning if the metadata for this dataset has already been set somewhere lower down in the heirarchy than the requested group. The result of this is that this function will not alter the resulting units for the dataset because it will be overshadowed by the metadata setting further down. (see warnings for an explanation of warnings)
- irate.core.get_units(dataset)¶
Gets the units for a dataset in an IRATE file.
Because units are typically expressed in factors of the hubble constant and may or may not be comoving or otherwise varying with scale factor, two additional factors are needed beyond the raw unit conversion. To use these together, the appropriate factor to multiple the dataset by to get to physical units for an assumed hubble parameter h and scale factor a is: tocgs`*h^`hubbleexponent`*a^`scalefactorexponent . Hence, for example, comoving Mpc/h would have tocgs =3.0857e24 , hubbleexponent =-1, and scalefactorexponent =1 .
Parameters: dataset – A Dataset object in an IRATE file Returns: name,tocgs,hubbleexponent,scalefactorexponent. name is the name of the unit for this dataset, tocgs is a factor to multiply the dataset by to get cgs units, hubleexponent is the exponent of the reduced hubble parameter (h=H0/100) for this unit, and scalefactorexponent is the exponent describing how this unit varies with the scale factor. Alternatively, if no unit is found, None is returned. Note
I f you are not working with a file where you know for sure what the units are, is important to check whether or not the unit is None, because the IRATE format does not require units for all datasets, as some quantities are dimensionless. The easiest way to do this is:
res = get_units(mydataset) if res is None: ... do something if it is unitless ... else: nm,tocgs,hexp,sfexp = res ... do something with the units ...
Raises TypeError: If dataset is not a dataset
- irate.core.set_units(dataset, unitname, tocgs, hubbleexponent, scalefactorexponent, unitgroup=None)¶
Sets the units of a dataset to the provided values.
Because units are typically expressed in factors of the hubble constant and may or may not be comoving or otherwise varying with scale factor, two additional factors are needed beyond the raw unit conversion. To use these together, the appropriate factor to multiple the dataset by to get to physical units for an assumed hubble parameter h and scale factor a is: tocgs`*h^`hubbleexponent`*a^`scalefactorexponent . Hence, for example, comoving Mpc/h would have tocgs =3.0857e24 , hubbleexponent =-1, and scalefactorexponent =1 .
Parameters: - dataset – The dataset for which to set the units.
- unitname (str) – A human-readable name of the unit.
- tocgs (str) – A numerical factor to multiply the dataset by to convert to cgs units.
- hubbleexponent (str) – The exponent of the hubble parameter for this unit.
- scalefactorexponent (str) – The exponent of the scale factor for this unit.
Raises TypeError: If an input is not the correct type
- irate.core.get_cgs_factor(dataset, h=0.7, a=1)¶
Retrieves the factor to multiply by the dataset to convert to physical CGS units at a given scale factor and hubble constant.
Parameters: - dataset – An Dataset for which to read the units.
- h – Reduced hubble parameter to assume for the conversion.
- a – Scale factor to assume for the conversion = 1/(z+1).
Raises: - TypeError – If dataset is not a dataset
- ValueError – If the dataset is dimensionless
irate.validate Module¶
The irate.validate module contains classes and functions for testing if a file conforms to the IRATE specification. It also contains tools for easily extending the validator to allow 3rd parties to provide validator modules for their formats.
See Custom Validators/Extending the IRATE Format for more details on how to write custom extensions to the IRATE format.
- exception irate.validate.IRATEFormatError(msg, h5grpname=None, validator=None)¶
This exception is raised if a file is loaded that does not conform to the IRATE Format.
- class irate.validate.RootValidator(parentvalidator, printmsg=None, immediatefail=None)¶
Validates the root level of an IRATE file.
- class irate.validate.Validator(parentvalidator, printmsg=None, immediatefail=None)¶
The base class for classes that are intended to validate IRATE format files.
To implement a validator, a subclass must override validate(), which will be valled to validate a Group. If not, an object of the subclass will not be able to be created, as this is an abstract class. See the Validator.validate docstring for the correct way to report format errors when they are encountered.
A subclass should also define a groupname attribute at the class level - that name will be used to identify groups that are to be assigned to that validator.
- check_units(dataset, unitname=None, invalidate=True)¶
Check the units on a dataset
Parameters: Returns: True if the unit check succeeded, False if it failed because the name of the units does not match unitname, or None if it failed because no units are present.
Note
If the dataset provided is not a dataset, this method will call invalid() with a message indicating that this occured.
- validate(grp)¶
Subclasses should override this - it is called to validate a particular h5py.Group that is matched to this validator.
If a problem is encountered, the validator should call Validator.invalid() with a message describing the problem. If a dataset on the validated group should have units, the check_units() method should be called on the dataset to check for units (or a specific unit name, if desired)
After this method finishes, all subgroups will also be validated - to skip validation for some subgroups, set the skipsubgroups attribute of this object to either a list of names to skip, or True to not validate any subgroups.
Thus, a simple example might be:
def validate(self,grp): if 'somegroup' not in grp: self.invalid('somegroup missing!') self.skipsubgroups = ['ignorethisgroupname'] self.check_units(grp['datasetwunits']) self.check_units(grp['distance'],'kpc')
- irate.validate.activate_validator_type(tnames, defaultlast=True)¶
Sets the active list of validators to those for the requested type.
Parameters: - tnames – The name of the type (group of validators) to activate or a list of types to activiate. Note that the order of types determines the order in which validators are checked for matching a particular group in an IRATE file.
- defaultlast – If True, when a list of types is given and the default validators are present, the default validators will be set to run only after all other validators have been run.
Raises KeyError: If tname is not a validator type name.
- irate.validate.add_validator_type(tname, validators, includedefaults=True)¶
Adds a set of validator classes as a type.
Parameters: - tname – The name for this type (group of validators).
- validators – A sequence of subclasses of Validator to use as validators for this type.
- includedefaults (bool) – If True, the default validators are added after the supplied validators. This is usually what you want because otherwise there’s no guarantee the file will actually follow the IRATE format.
Raises TypeError: If something in validators is not a Validator subclass.
- irate.validate.find_custom_validator_dir()¶
Identifies and returns the directory that stores custom validators.
Returns: The directory where validator files are supposed to live Raises OSError: If no suitable directory can be found.
- irate.validate.find_validator(groupname)¶
Locates the appropriate validator based on the given group name.
Parameters: groupname – The name of the group to match against a Validator groupname. Returns: The class matched to the provided group name or False if the search failed.
- irate.validate.get_validator_types(tname=None)¶
Returns a list of all the validator types available, or all the validator classes associated with a given validator type.
Parameters: tname – If None, returns a list of validator types, or if a validator type, returns a list of Validator subclasses for that type. Returns: A list of types if tname is None or a list of Validator subclasses Raises KeyError: If tname is not a validator type name.
- irate.validate.register_validator(valcls, front=True)¶
Registers a Validator class for use by the find_validator() function.
Parameters: - valcls – A class that is a subclass of Validator. The matching group names will be inferred from its groupname attribute.
- front – If True, the given validator will be registered in front of others - e.g. a group that matches its name will not move on to the others. Otherwise it will be placed in the back.
- irate.validate.remove_validator_type(tname)¶
Removes a set of type validators. :param tname: The name of the type to have its validators removed.
Raises KeyError: If tname is not a validator type name.
- irate.validate.validate_file(fnorfile, verbose=False, immediatefail=False)¶
Tests whether or not the file conforms to the IRATE format.
Parameters: - fnorfile – The filename or h5py.File to test.
- verbose (bool) – If True, informational messages will be printed to stdout, and if False no messages will be printed. Can also be a callable f(str) that will be called each time a message is to be printed.
- immediatefail (bool) – If True, an exception is raise when the first invalid aspect of the file is encountered. Otherwise, the function completes and returns all errors.
Returns: A list of errors encountered (or an empty list if valid).
Raises IRATEFormatError: If any part of the file does not conform to the standard and immediatefail is True.
irate.ahf Module¶
- irate.ahf.ahf_halos(fname, outname, snapnum, nogas=False, name='HaloCatalog_AHF', posname='comoving Mpc/h', posunit=array([ 3.08568025e+24, -1.00000000e+00, 1.00000000e+00]), velname='km/s', velunit=array([ 100000., 0., 0.]), mname='M_sun/h', munit=array([ 1.98892000e+33, -1.00000000e+00, 0.00000000e+00]), radname='comoving kpc/h', radunit=array([ 3.08568025e+21, -1.00000000e+00, 1.00000000e+00]), enname='M_sun/h*(km/s)^2', enunit=array([ 1.98892000e+43, -1.00000000e+00, 0.00000000e+00]), phiname='(km/s)^2', phiunit=array([ 1.00000000e+10, 0.00000000e+00, 0.00000000e+00]))¶
Reads an AHF_halos file, then collimates it such that a list of column headers and a list of arrays, each of which corresponds to a column. All entries in the columns will be in the same order–i.e. the ith entry in column x corresponds to the same halo as the ith entry in column y.
Parameters: - fname – The file to be read; should be in ASCII format with column headers preceeded by a #
- outname – The IRATE format file to either be created or to have data added to it, if it already exists.
- snapnum (int) – Specifies the snapshot number that the data will be saved under in the IRATE file; that is, the data ends up in the group “Snapshot”+snapnum
- nogas (bool) – True if AHF was NOT compiled with -DGAS_PARTICLES; otherwise this will fail in attempting to collimate the data because it will try to combine columns (e.g. Lx_gas, Ly_gas, and Lz_gas) that don’t exist.
- name – Specifies the name of the group that holds the datasets. Data is saved under “Snapshot”+snapnum+”/”+name
- ____name – A human readable string that identifies the units used for the property specified by ____ (position, velocity, mass, radius, energy, and phi0)
- ____units – A three element array that gives, in order, the conversion factor between the units used for the property given by * to CGS, the exponent on the reduced Hubble Parameter that appears in that unit, and the exponent on the scale factor that appears in that unit.
Returns: An array that contains the number of bins used for each halo in the radial profiles file, for the purpose of reading the .AHF_profiles file.
- irate.ahf.ahf_param(pfile, outname, snapnum, name='HaloCatalog_AHF')¶
Reads an AHF .parameter file and adds it to an existing IRATE file as attributes to the /CatalogHeader/AHF group.
Parameters: - pfile – The parameter file to be read. Should be standard AHF ASCII parameter file format.
- outname – The IRATE file to store the parameters in.
- irate.ahf.ahf_particles(fname, outname, snapnum, pfilename=None, name='HaloCatalog_AHF')¶
Read a .AHF_particles file and save an array with all the particles in it in order, along with a list of the number of particles in each halo to an IRATE file. If there’s a particle identifier, the array is Nx2 with the identifier in the second column.
Parameters: - fname – The .AHF_particles file to be read. Must be in ASCII format
- outname – The IRATE file that the data is linked to
- pfilename – The HDF5 file that the data is saved in
- snapnum – The snapshot number that identifies the group that the halo catalog belongs to in the IRATE file
- name – The name of the halo catalog in the IRATE file that the particles correlate to.
- irate.ahf.ahf_profiles(fname, nhalos, nbins, outname, snapnum, name='HaloCatalog_AHF', radname='comoving kpc/h', radunits=array([ 3.08568025e+21, -1.00000000e+00, 1.00000000e+00]), mname='M_sun/h', munits=array([ 1.98892000e+33, -1.00000000e+00, 0.00000000e+00]), velname='km/s', velunits=array([ 100000., 0., 0.]), angname='(M_sun/h)*(Mpc/h)*(km/s)', angunits=array([ 6.13717116e+62, -2.00000000e+00, 1.00000000e+00]), enname='M_sun/h*(km/s)^2', enunits=array([ 1.98892000e+43, -1.00000000e+00, 0.00000000e+00]), nogas=False)¶
Read an ASCII .AHF_profiles file, collimate it, and return a list of data and a list of column headers. The list of data is a list over columns, then each halo has it’s own list within the columns.
Parameters: - fname – The ASCII format .AHF_profiles file to be read.
- nhalos (int) – The number of halos contained in the file
- nbins – An array or list that tells you the number of radial bins for each halo
- outname – The existing IRATE file to save radial profile data to
- snapnum (int) – The number of the snapshot that the halo catalog that this data belongs to is under in the existing IRATE file
- name – The name of the group that contains the halo catalog
- ____name – A human readable string that gives the units for the given property: radius, mass, velocity, angular momentum, or energy
- ____units – A three element array, the first of which tells the conversion factor between the units of * and CGS, the second of which gives the exponent on h as it appears in those units, and the third gives the exponent on a as it appears in those units.
- nogas (bool) – True if AHF was NOT compiled with -DGAS_PARTICLES; otherwise this will fail in attempting to collimate the data because it will try to combine columns (e.g. Lx_gas, Ly_gas, and Lz_gas) that don’t exist.
- irate.ahf.read_and_write_particles(infile, outfile, maxsize)¶
The HDF5 v 1.8 method of reading and writing particles should allow me to use a lot less memory by periodically writing to the file (since v 1.8 allows for appending to datasets). Note that this will probably be slower than the 1.6 method, at least for small files, but if memory is a problem, this is probably the way to go.
Warning
THIS DOESN’T WORK RIGHT NOW. I can’t get the appending to work, so if you desperately want to use this function, you’ll have to fix it. In fact, I should probably just delete it, but I always hate doing that Furthermore, since it isn’t working, it hasn’t been updated to work adhere to the new IRATE format, so this function absolutely does NOT work.
Parameters: - infile – The AHF_particles file to read. Must be in ASCII format.
- outfile – The IRATE catalog file to save the particle data to.
- maxsize – The maximum amount of memory that can be used at any given time, in GB.
irate.gadget Module¶
- irate.gadget.gadget_hdf5_to_irate(inname, outname, snapnum, t0_name='Gas', t1_name='Dark_Halo', t2_name='Dark_Disk', t3_name='Dark_Bulge', t4_name='Star', t5_name='Dark_Boundary', s8=None, ns=None, omegaB=None, lname='comoving Mpc/h', lunits=array([ 3.08568025e+24, -1.00000000e+00, 1.00000000e+00]), vname='(km/s)*sqrt(a)', vunits=array([ 1.00000000e+05, 0.00000000e+00, 5.00000000e-01]), mname='1e10 M_sun/h', munits=array([ 1.98892000e+43, -1.00000000e+00, 0.00000000e+00]))¶
Transforms Gadget type 3 (HDF5) snapshots into a format that meets the IRATE specifications. This will automatically check for Makefile enabled blocks.
Parameters: - inname – Gadget type 3 snapshot to convert to IRATE
- outname – IRATE file to output
- snapnum (int) – The number to add to ‘Snapshot’ that becomes the name of the group that particle data is added to.
- #name – Name of group that particles of type # are given
- s8 (float) – sigma_8, for the purposes of adding it to the Cosmology group
- ns (float) – n_s, for the purposes of adding it to the Cosmology group
- omegaB (float) – omegaB, for the purposes of adding it to the Cosmology group
- ___name – A human readable string that defines the units for either length (l), velocity (v), or mass (m)
- ___units – A three-element array. The first entry defines the conversion factor to convert either length (l), velocity (v), or mass (m) to CGS units. The second element is the exponent on the reduced Hubble Parameter that appears in that unit, and the third is the exponent on the scale factor that appears in that unit.
# refers to the same names as Gadget2:
- 0 = gas particles
- 1 = halo particles
- 2 = disk particles
- 3 = bulge particles
- 4 = star particles
- 5 = bndry particles
- irate.gadget.gbin2_to_irate(inname, outname, snapnum, t0_name='Gas', t1_name='Dark_Halo', t2_name='Dark_Disk', t3_name='Dark_Bulge', t4_name='Star', t5_name='Dark_Boundary', s8=None, ns=None, omegaB=None, lname='comoving Mpc/h', lunits=array([ 3.08568025e+24, -1.00000000e+00, 1.00000000e+00]), vname='(km/s)*sqrt(a)', vunits=array([ 1.00000000e+05, 0.00000000e+00, 5.00000000e-01]), mname='1e10 M_sun/h', munits=array([ 1.98892000e+43, -1.00000000e+00, 0.00000000e+00]))¶
Reads a GADGET type 2 snapshot file block by block (e.g. coordinate block for gas particles), writes the block to an IRATE formate HDF5 file, and then deletes that block from memory. If a given block identifier isn’t recognized, one of two things will happen: either the script will figure out which particles that data belongs to and add it automatically with the dataset named according to the label used, or if it’s not obvious what particles the data belongs to, the user will be given the option to skip it or to exit entirely.
Parameters: - inname – The name of the gadget binary file to be read
- outname – The name of the IRATE file to be written,
- snapnum (int) – The number to add to ‘Snapshot’ that becomes the name of the group that particle data is added to.
- t#_name – Determines the name of the group that contains the data from #
- s8 (float) – sigma_8, for the purposes of adding it to the Cosmology group
- ns (float) – n_s, for the purposes of adding it to the Cosmology group
- omegaB (float) – omegaB, for the purposes of adding it to the Cosmology group
- ___name – A human readable string that defines the units for either length (l), velocity (v), or mass (m)
- ___units – A three-element array. The first entry defines the conversion factor to convert either length (l), velocity (v), or mass (m) to CGS units. The second element is the exponent on the reduced Hubble Parameter that appears in that unit, and the third is the exponent on the scale factor that appears in that unit.
# refers to the same names as Gadget2:
- 0 = gas particles
- 1 = halo particles
- 2 = disk particles
- 3 = bulge particles
- 4 = star particles
- 5 = bndry particles
- irate.gadget.gbin_to_irate(inname, outname, snapnum, potential=False, accel=False, entropy=False, timestep=False, t0_name='Gas', t1_name='Dark_Halo', t2_name='Dark_Disk', t3_name='Dark_Bulge', t4_name='Star', t5_name='Dark_Boundary', ics=False, s8=None, ns=None, omegaB=None, lname='comoving Mpc/h', lunits=array([ 3.08568025e+24, -1.00000000e+00, 1.00000000e+00]), vname='(km/s)*sqrt(a)', vunits=array([ 1.00000000e+05, 0.00000000e+00, 5.00000000e-01]), mname='1e10 M_sun/h', munits=array([ 1.98892000e+43, -1.00000000e+00, 0.00000000e+00]))¶
Reads a GADGET format file block by block (e.g. coordinate block for gas particles), writes the block to an IRATE formate HDF5 file, and then deletes that block from memory.
Parameters: - inname – The name of the gadget binary file to be read
- outname – The name of the IRATE file to be written
- snapnum (int) – The number to add to ‘Snapshot’ that becomes the name of the group that particle data is added to.
- potential (bool) – True to read the Makefile enabled potential block
- accel (bool) – True to read the Makefile enabled acceleration block
- entropy (bool) – True to read the Makefile enabled dA/dt block
- timestep (bool) – True to read the Makefile enabled timestep block
- t#_name – Determines the name of the group that contains the data from #
- ics (bool) – True if the file being converted is an initial conditions file, in which case the gas density and smoothing length blocks won’t be looked for.
- s8 (float) – sigma_8, for the purposes of adding it to the Cosmology group
- ns (float) – n_s, for the purposes of adding it to the Cosmology group
- ___name – A human readable string that defines the units for either length (l), velocity (v), or mass (m)
- ___units – A three-element array. The first entry defines the conversion factor to convert either length (l), velocity (v), or mass (m) to CGS units. The second element is the exponent on the reduced Hubble Parameter that appears in that unit, and the third is the exponent on the scale factor that appears in that unit.
# refers to the same names as Gadget2:
- 0 = gas particles
- 1 = halo particles
- 2 = disk particles
- 3 = bulge particles
- 4 = star particles
- 5 = bndry particles
- irate.gadget.irate_to_gbin(inname, outname, snapnum)¶
Converts a single snapshot in an IRATE format file into a SnapFormat = 1 Gadget binary file. If there are groups with “Star” in the name, they’re assumed to contain star particles and will be placed in Gadget group 4; likewise, if there are groups with “Gas” in the name, their particles will be placed in Gadget group 0. All other particles will be placed in Gadget group 5.
Parameters: - inname – The input IRATE format file.
- outname – The name of the output Gadget file
- snapnum (int) – The integer identifying where the data is stored; that is, particle data is expected to be under ‘/Snapshot{snapnum}/ParticleData.
- irate.gadget.read_gbin(fname, potential, accel, entropy, timestep)¶
- Reads a GADGET format file and returns the data in lists, one for
- each type of particle that contains arrays for each bit of data about each particle.
Warning
Currently unused in the IRATE suite, and definitely not maintained either. Use at your own risk.
Parameters: - fname – The name of the gadget binary file to be read
- potential (bool) – True to read the Makefile enabled potential block
- accel (bool) – True to read the Makefile enabled acceleration block
- entropy (bool) – True to read the Makefile enabled dA/dt block
- timestep (bool) – True to read the Makefile enabled timestep block
irate.enbid Module¶
- irate.enbid.add_enbid(outname, header, gasrho, halorho, diskrho, bulgerho, starrho, bndryrho, gastree, gasname, halotree, haloname, disktree, diskname, bulgetree, bulgename, startree, starname, bndrytree, bndryname)¶
Writes the already read data from EnBiD to an existing IRATE file under the Analysis group.
Parameters: - outname – The IRATE file to save the data to. Must exist already and must already have a /Analysis group (and not have a /Analysis/EnBiD group
- header – The EnBiD header, in the order that it’s provided (which is the same as it’s provided in Gadget files.
- ___rho – The density arrays for each group of files to be saved.
- ___tree – The group to save the data for each respective particle type under. Must be either dark, star, or gas.
- ___name – The name to give the datasets for each respective particle type.
Warning
This function hasn’t been maintained and updated to the new format yet, so it’s not yet ready for use.
- irate.enbid.read_enbid_binary(fname)¶
- Reads a binary file outputted by EnBiD with ICFormat = 1 – that is,
- with a Gadget format header and data in binary format, written as float data, in the same order as the Gadget file that EnBiD was run on. Returns the header, then the data for each group as separate arrays.
Parameters: fname – The binary file to read. Warning
This function hasn’t been maintained and updated to the new format yet, so it’s not yet ready for use.
irate.rockstar Module¶
- irate.rockstar.rockstar_ascii(fname, outname, snapnum, name='HaloCatalog_Rockstar', s8=None, ns=None, omegaB=None)¶
Reads a Rockstar ASCII file and collimates it, then saves the data in an IRATE format file (creating one if it doesn’t exist).
Parameters: - fname – The input ASCII file to read and collimate.
- outname – The IRATE file to save the data in
- snapnum – The integer snapshot number, which the data will be saved under in the IRATE file.
- name – The name of the group that will contain the datasets that hold the data
- s8 – sigma_8, for inclusion in the cosmology group. May be None.
- ns – Power Spectrum Index, for inclusion in the cosmology group. May be None.
irate.tipsy Module¶
- irate.tipsy.load_tipsy_binary()¶
load_tipsy_binary(fn, verbose=True, unitconv=True, compression=None, printiters=100000)
Load a Tipsy binary file using the tipsyapi tools.
Parameters: - fn (str) – file name or ‘stdin’ to use stdin
- verbose (bool) – If True, information is printed as the file is loaded
- unitconv (bool) – If True, converts from tipsy/PKDGRAV system units to units where velocities are km/s, positions are in kpc, and masses are in Msun (although note that the gravitational potential is left in Tipsy units e.g. G=1). Alternatively, it can be a dictionary mapping names of fields e.g. (‘Velocity’,’Position’, and ‘Mass’) to a scaling factor that the input data will be multiplied by to give output data. If None, no unit scaling will be applied.
- printiters (int) – The number of iterations to wait before printing information about the current particle. e.g. 1 will print info for every particle. If <=0, nothing will be printed.
Returns: A dictionary with ‘dark’,’star’, and ‘gas’ entries that are dictionaries of strings to arrays that match the IRATE format.
- irate.tipsy.tipsy_to_hdf5()¶
tipsy_to_hdf5(tipsyfn, hdfn=None, verbose=True, unitconv=True, compression=None, printiters=100000)
Converts a tipsy file to an equivalent HDF5 file.
Parameters: - tipsyfn (str) – The file name of the tipsy file to read.
- hdfn (str or None) – The file name of the hdf5 file to write (will be appended with .h5 if not present). If None, the written file will be the same as tipsyfn with ‘.h5’ appended.
- compression – The compression type to use for the datasets or None for no compression.
Other arguments are the same as load_tipsy_binary().
Returns: The h5py.File object (not closed).